ABOUT US
Our lab studies fundamental epigenetic pathways and the mechanisms through which these processes are corrupted by disease-associated mutations in chromatin regulators. We aim to (1) understand the molecular mechanisms utilised by chromatin regulators to exert their function in healthy human cells, and (2) examine how these mechanisms are altered in human disorders on a molecular level.
The advent of next-generation sequencing and the discovery of CRISPR has allowed for genetic studies to be performed in virtually any cell type. We take advantage of existing high-throughput technologies, such as CRISPR/Cas9 forward genetic screens, to discover novel factors involved in chromatin regulation. To examine the role of these factors in the pathway of interest, we then use a combination of genetic and biochemical approaches. We are also looking to expand the genetic toolbox we have available and so we develop novel genetic methods to study epigenetic pathways in human cells. Some examples of the type of approaches we use can be found below:
- Genetically encoded fluorescent reporters
- CRISPR/Cas9 forward genetic screens
- CRISPRi and CRISPRa
- Genome editing
- Next-generation sequencing
- Flow cytometry
- Immunofluorescence microscopy
- Chromatin immunoprecipitation (ChIP)
The HUSH complex
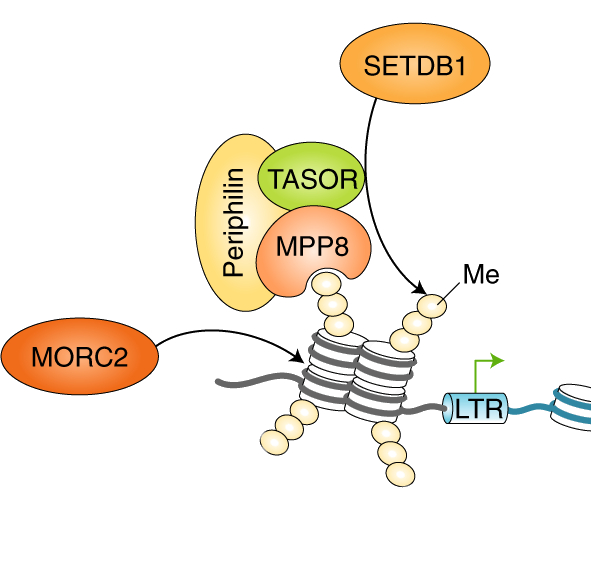
A major focus of work in the lab is characterising epigenetic repression by the HUSH complex, the key mediator of position-effect variegation in human cells. Iva discovered the HUSH complex during her PhD through a series of haploid and CRISPR/Cas9 genetic screens. HUSH localises to genomic loci rich in the repressive histone modification H3K9me3, where subsequent recruitment of the methyltransferase SETDB1 and the chromatin remodeller MORC2 is required for H3K9me3 deposition and chromatin compaction to instil epigenetic repression. Independent studies have shown a role for HUSH in the silencing of retroelements and unintegrated retroviral DNA, and as a restriction factor for HIV. Achieving a mechanistic understanding of HUSH-mediated repression is a major focus of our ongoing research.
Chromatin state reporters
While a functional reporter assay was the critical factor that enabled us to identify and characterise the HUSH complex, no analogous reporters currently exist to allow a similar genetic interrogation of other major epigenetic complexes. Therefore, a major focus of work in the lab is the generation of cell lines harbouring fluorescent transgenes that report on the chromatin environment in which they are situated. These phenotypic reporters can be subsequently interrogated through a battery of genetic approaches to gain new insight into the underlying biology of any epigenetic pathway.

Cancer epigenetics
As many as half of human cancers contain mutations in epigenetic modifiers. In order to understand how epigenetic pathways are dysregulated upon malignant transformation, we first need to have a detailed understanding of the underlying pathways in healthy cells. To this end, we harness the power of genetic approaches in cell lines harbouring fluorescent reporters for chromatin pathways that are frequently dysregulated in cancer, with a particular focus on histone methylation. Crucially, we can also use such chromatin reporter lines to perform chemical screens and identify small molecule inhibitors for these different pathways.

Lineage specification
While cell tyme-specific silencing of lineage-inappropriate genes has been classically attributed to the repressive H3K27me3 histone modification, recent findings indicate that H3K9me3, best known for its role in silencing repetitive elements and non-coding parts of the genome, plays an important role in controlling cell identity. We are interested in the molecular mechanisms of H3K9me3-mediated silencing of lineage-inappropriate genes, with a particular focus on the recently discovered HUSH complex.

Technology development
We exploit a wide variety of genetic techniques to study chromatin biology. In addition to using established methodologies such as CRISPR/Cas9-mediated genetic screens, an important focus of our work is the development of novel genetic approaches to study chromatin-mediated effects on gene expression. One recent example is the development of DIVA, a genome-wide method which uses the large-scale mapping of retroviral integration sites to probe chromatin accessibility.
